|

Predicting the value of free energy
change
A test case
Suppose that the task is predicting the protein stability change when
Glu (E) 200 is mutated into Lys (K) in the Prion Protein at pH 7
and Temperature equal to 5 °C (Fig.2) and that the sequence is
available. This can be done either with proteins that are already known
with atomic resolution or with any protein sequence
available.
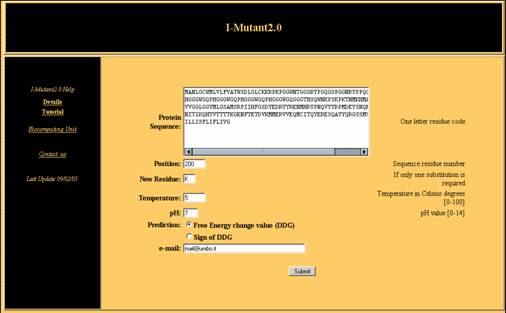
Input
The following operations are required:
-
Fill in the Protein
Sequence box with the one letter code sequence of your protein.
-
In the Position
box insert the number of the residue that undergoes mutation.
- In New Residue insert
the required one letter code mutation (optional).
-
The boxes pH
and Temperature can be filled with the experimental
conditions at which the experiment is simulated.
The selection of the
radio button “Free Energy change value (DDG)”
allows the prediction of
the free energy change value. The output of the predictor will be send
to your e-mail to address, if required, or shown directly to you in due
time.
The following operations are required:
-
Fill in the Protein
Sequence box with the one letter code sequence of your
protein.
-
In the Position
box insert the PDB number of the residue that undergoes mutation.
-
The boxes pH
and Temperature can be filled with the experimental
conditions at which the experiment is simulated.
The selection of the radio button “Free
Energy change value (DDG)” allows the
prediction of the free energy change value. The output of the predictor
will be send to the your e-mail to address, if required or given
directly to you.
Output
The results obtained for the given test case are listed below:
SEQ File: /junk/I-Mutant/Mut17864/fileseq.txt
Position WT NEW DDG pH T
200 E V 0.75 7.0 5
200 E L 0.86 7.0 5
200 E I 1.31 7.0 5
200 E M 0.93 7.0 5
200 E F 1.99 7.0 5
200 E W 0.47 7.0 5
200 E Y 1.09 7.0 5
200 E G -0.61 7.0 5
200 E A -0.30 7.0 5
200 E P -0.05 7.0 5
200 E S 0.76 7.0 5
200 E T 0.59 7.0 5
200 E C 0.15 7.0 5
200 E H -0.06 7.0 5
200 E R 1.18 7.0 5
200 E K 0.10 7.0 5
200 E Q 0.13 7.0 5
200 E N -0.03 7.0 5
200 E D 0.19 7.0 5
WT: Aminoacid in Wild-Type Protein
NEW: New Aminoacid after Mutation
DDG: DG(NewProtein)-DG(WildType) in Kcal/mol
DDG<0: Decrease Stability
DDG>0: Increase Stability
T: Temperature in Celsius degrees
pH: -log[H+]
If you do not ask for a specific mutation in the position at hand, all
the possible mutations will be taken into consideration, including the
one that may be of specific interest. However if you are asking only
one mutation, by activating the "New Residue" option, only one
prediction will be returned. The free energy change corresponding to
the mutation of Glu (E) 200 to Lys (K) in Prion Protein at temperature
of 5 Celsius degree and pH 7 is 0.10 Kcal/mol and is highlighted in
red.
|